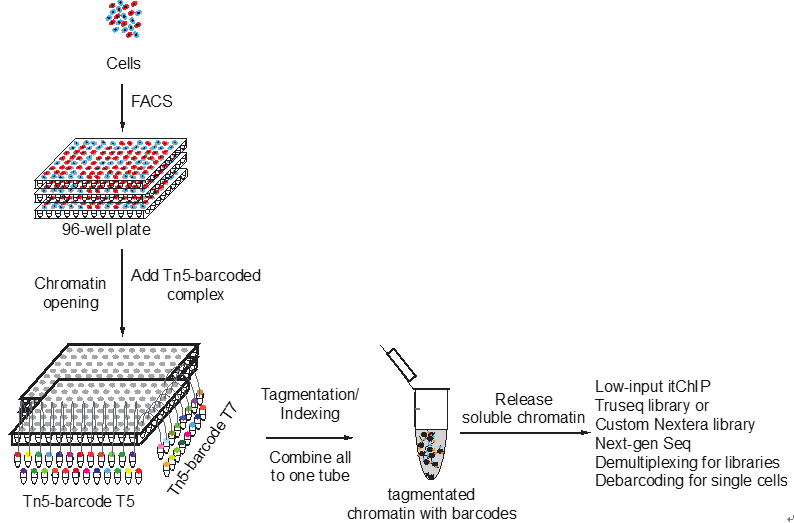
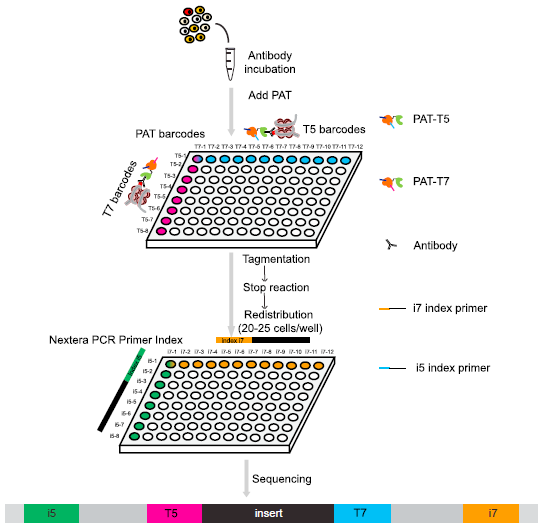
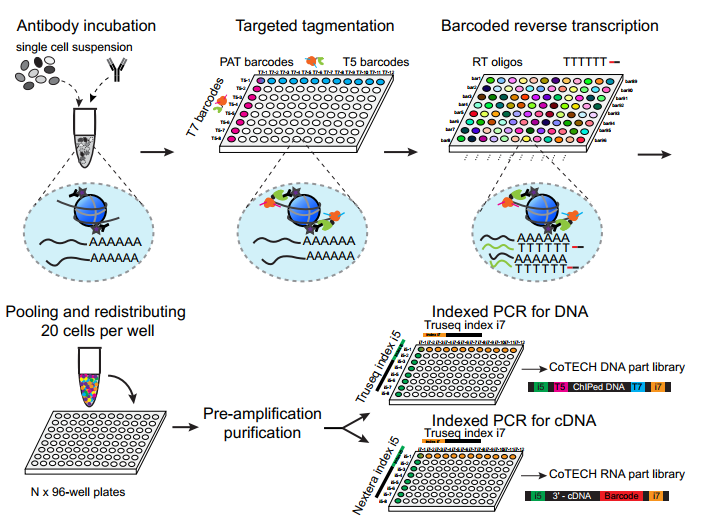
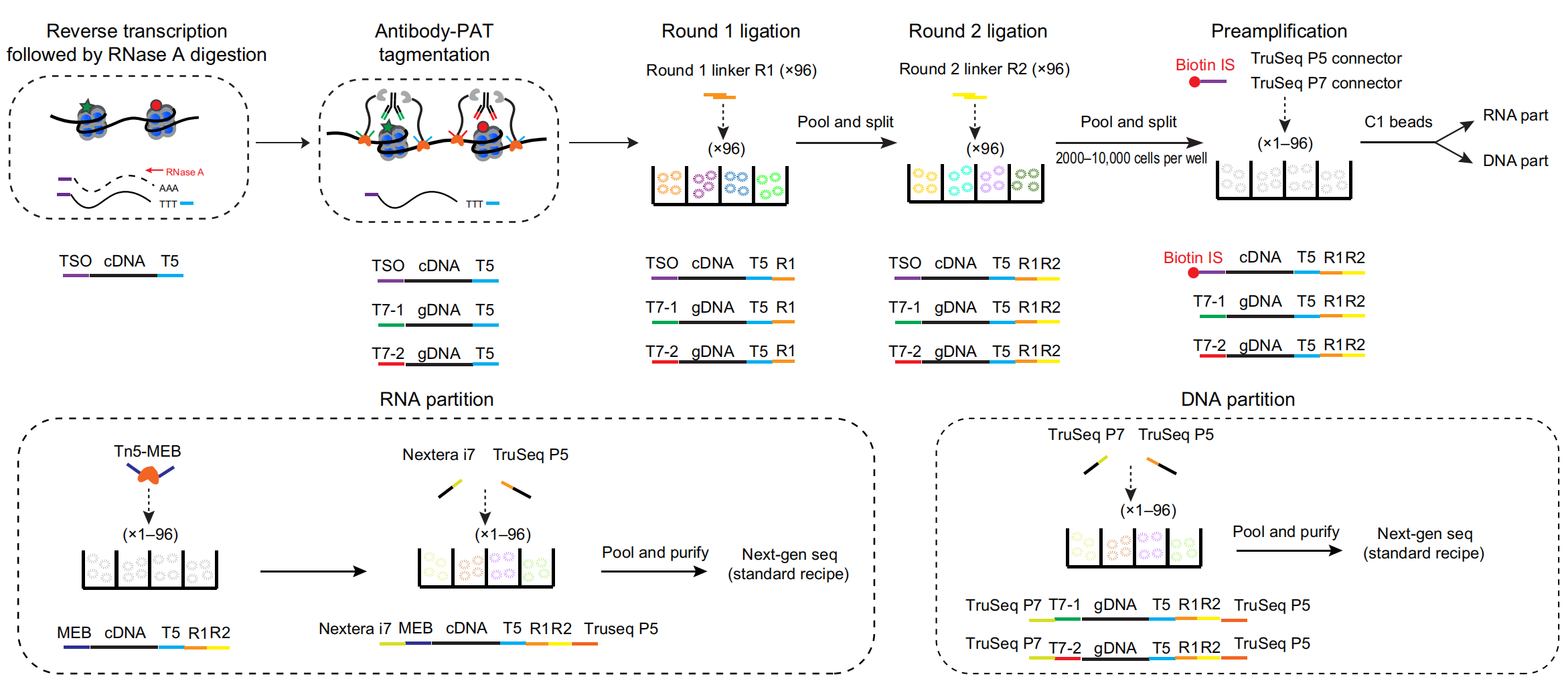
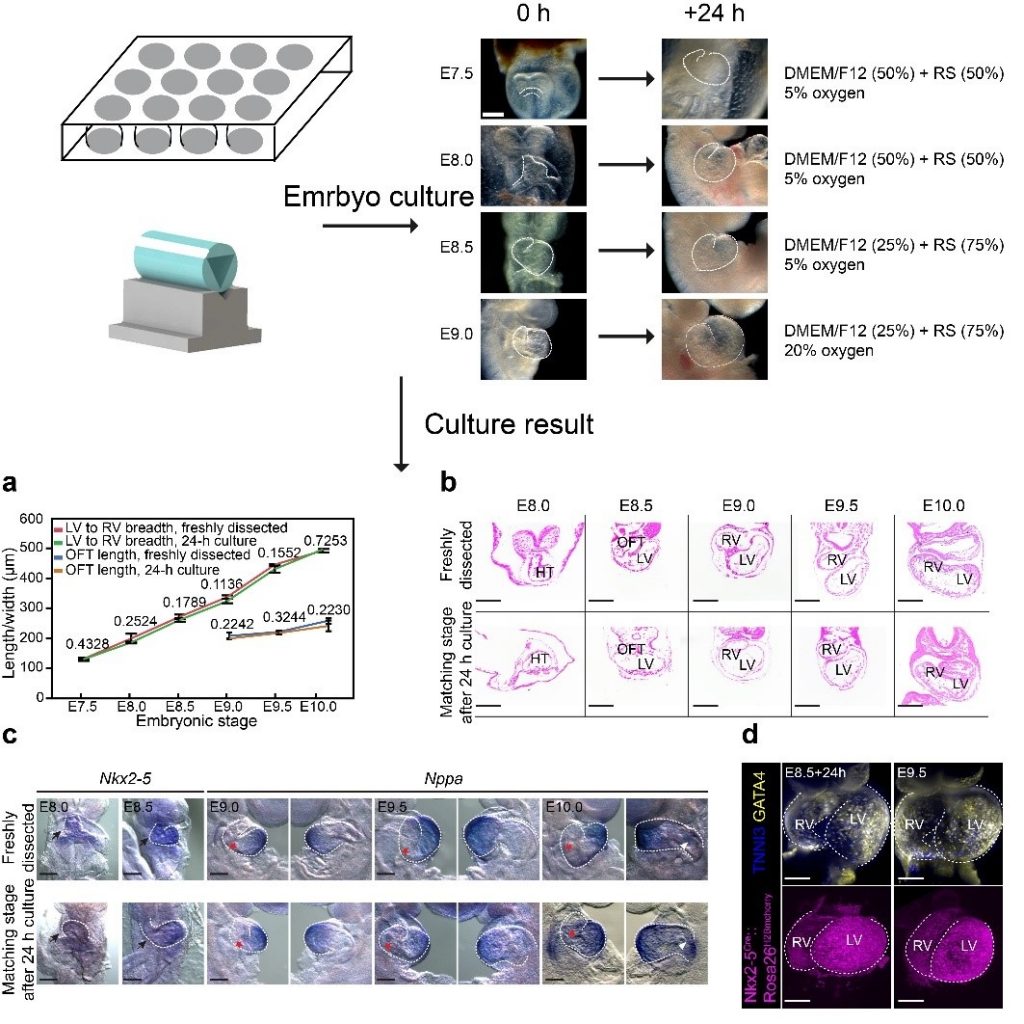
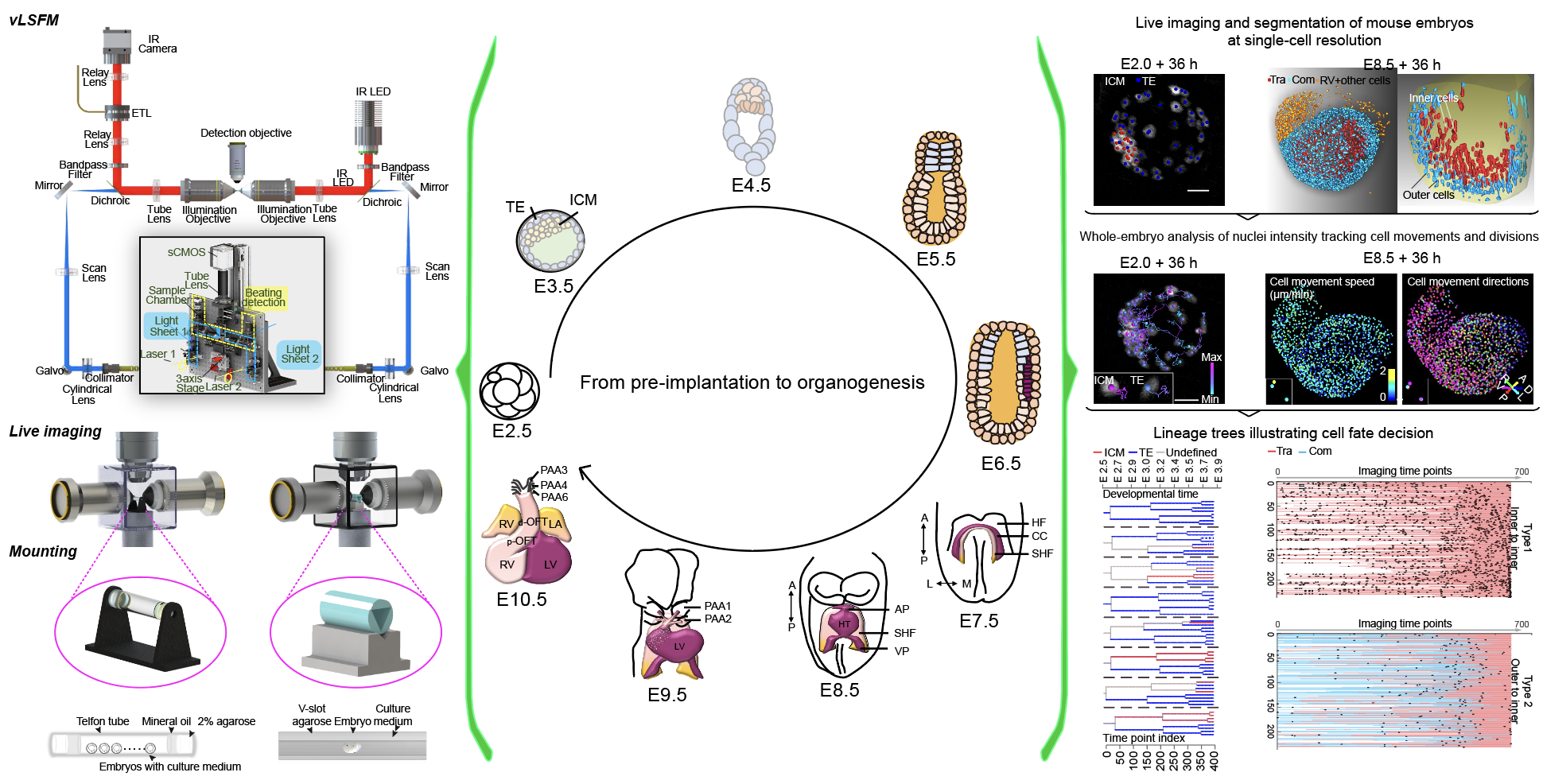
He laboratory is striving for developing and applying new technologies in single-cell omics and in toto live imaging for understanding of gene regulation and tissue-scale cell lineages. Next-generation sequencing (NGS) based multi-modal omics provides rich molecular characterizations with single-cell sensitivity but no cells can be assayed more than once in a destructive manner, while fluorescence microscopy imaging brings about comprehensive spatial and temporal information from morphological description of a tissue/organ building scheme to quantitative molecular and cellular profiling. Currently, the team exploits single-cell omics, such as CoBATCH, sc-itChIP-seq, CoTECH, uCoTarget and scEpichem, to reveal key links between multi-dimensional epigenome with varying chromatin modifications and transcription factors binding and gene transcription shaping cell fates in development and human diseases. Efforts are also made towards designing and applying large field of view and high-resolution imaging tools for live tracing of cellular behaviors, including lineage histories, patterning, division, migration and movement, sculpting embryonic tissue morphology in mouse models. More ambitiously, the He laboratory is integrating single-cell omics and live imaging to reconstruct a real-time cellular and molecular fate map of mouse gastrulation and cardiogenesis. Ultimately, they aim to apply in-house technologies to medical research, such as mechanisms in cardiovascular and blood diseases and diagnostic medicine.
 Links:
Links:
Peking University
College of Future Technology, Peking University
Peking-Tsinghua Center for life sciences
Postdoctoral positions are available!
Find us at the office
The He lab
Institute of Molecular Medicine,College of Future Technology, Peking University, Integrated Science Research Center, Equipment building #2, Rm 232-233 Beijing, China, 100871
HeLab位于中国北京市海淀区颐和园路5号北京大学燕园校区综合科研2号楼2楼232-233房间
Give us a ring
Phone: +86(10)-62744560
Email: ahe@pku.edu.cn