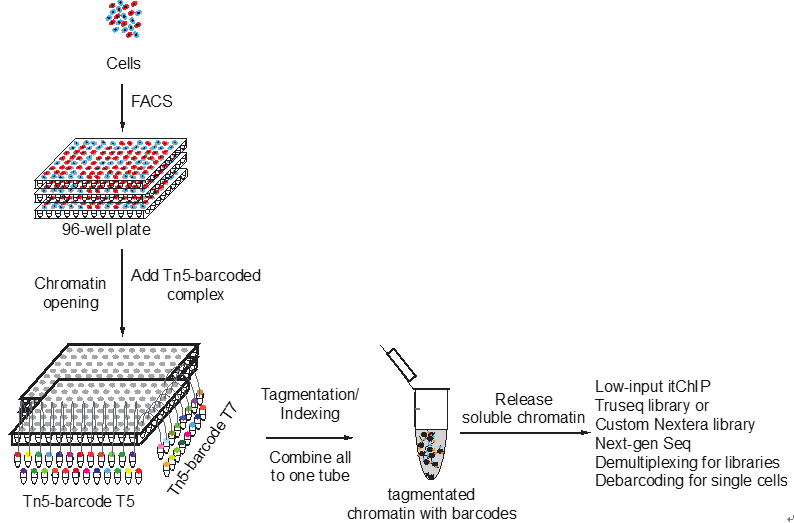
Single-cell itChIP
Single-cell simultaneous indexing and tagmentation-based ChIP-seq.
itChIP-seq is a low-cost and efficient method, compatible with both low cellular input and single cells for profiling chromatin states. itChIP combines chromatin opening, simultaneous cellular indexing and chromatin tagmentation within a single tube, enabling the processing of samples from tens of single cells to, more commonly, thousands of single cells per assay. We demonstrate that single-cell itChIP-seq (sc-itChIP-seq) yields ~9,000 unique reads per cell and is widely applicable and affordable for most biomedical research laboratories. (Ai et al. Nature Cell Biology, 2019)
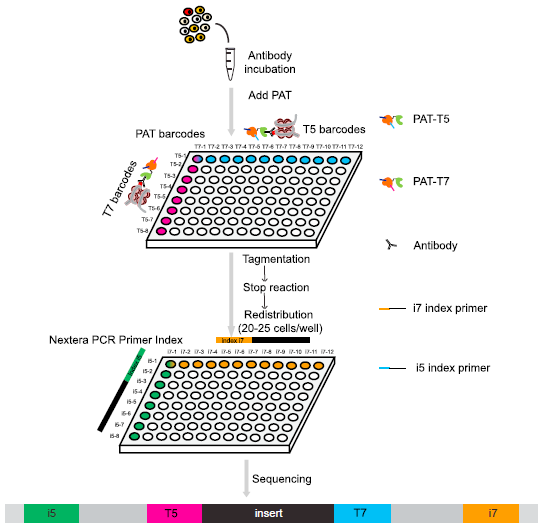
CoBATCH
Combinatorial barcoding and targeted chromatin release.
CoBATCH is a high-throughput single-cell ChIP-seq technology. Based on fusion protein-protein A-Tn5 (PAT), we develop in situ ChIP, which is suitable for as low as 100 initial cells and intact tissues. PAT is enriched through specific antibodies to genomic regions, and Tn5 generates indexed chromatin ready for library preparation and sequencing. By introducing combinatorial indexing strategy, we develop CoBATCH, a high efficient and high-throughput single-cell histone modification and DNA-binding protein profiling method, suitable for both native and fixed cell condition. CoBATCH produces ∼12,000 non-duplicated reads per cell with extremely low background. (Wang et al. Molecular Cell, 2019)
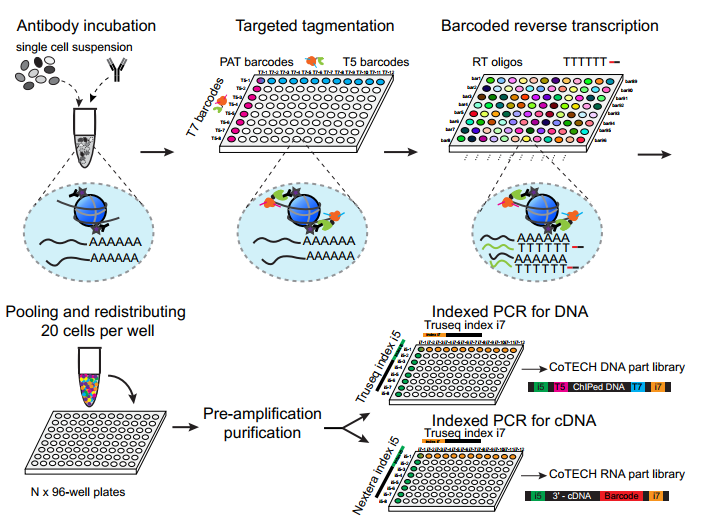
CoTECH
A joint method for profiling transcriptome and chromatin occupancy simultaneously in single cells.
We integrated our previously developed CoBATCH together with modified single-cell RNA-seq method, to simultaneously profile gene expression and chromatin binding in the same single cells. Furthermore, by aggregating data from cells with similar transcriptome (data from different experimental batches), we can obtain “pseudo-single-cells” that contain information on several histone modifications or TFs binding, enabling higher-dimensional epigenomic reconstruction. (Xiong et al. Nature Methods, 2021)
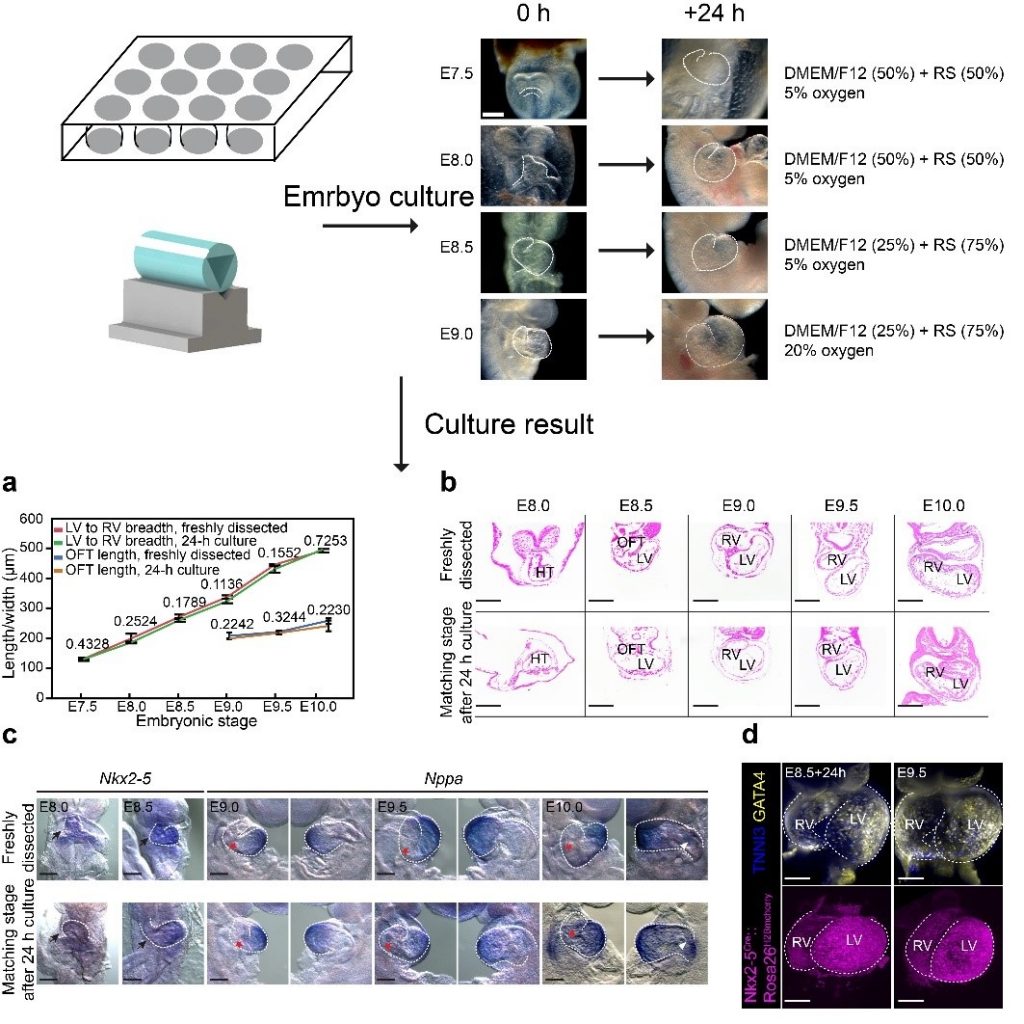
In vitro Embryo culture
We have established a highly effective embryo culture system for heart live-imaging.
We removed the yolk sac, EPC, and amnion of E8.25 embryos, while leaving them intact for embryos at E8.0 or earlier. Then embryos were cultured in the petri dish or V-Slot. Morphological, histological, and molecular analyses confirm that our culture conditions were able to support apparently normal heart development for more than 24 h with initiating stages from E6.5–E9.0, covering the developmental window from the early cardiac crescent stage (E7.5) to the early four-chambered stage (E10.0).
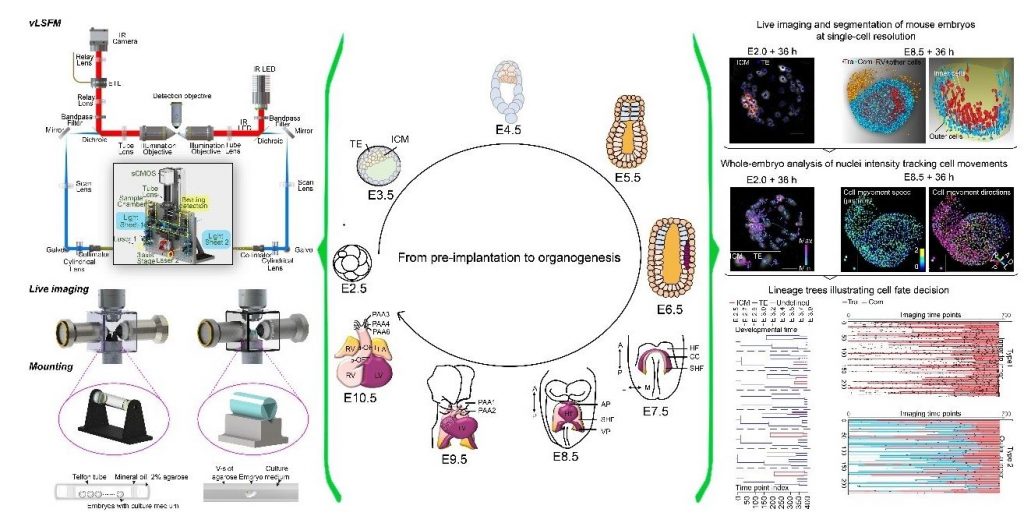
In toto live imaging
Time-resolved in toto cell lineage tracing.
Time-resolved in toto cell lineage tracing is fundamental for understanding the intercellular emergent properties for mammalian organogenesis. Having assembled an interdisciplinary group, we developed a customized vertical light-sheet microscope (vLSFM) equipped with an integrated embryo culture module, a heartbeat-gated imaging strategy, and a digital image processing framework for segmentation, tracking, and lineage tracing. Our integrated light-sheet microscopy-embryo culture system enables single-cell recording of pre-implantation to organogenesis. (Yue et al. Nature Cell Biology, 2020)(Zhang et al. Cell Stem Cell, 2023)
Video:
Video for In toto time-lapse imaging of mouse pre-implantation development
Video for Visualization and color indexing of cell division rates in pre-implantation development
Video for Inject beads into the cavity and subsequently directional movement
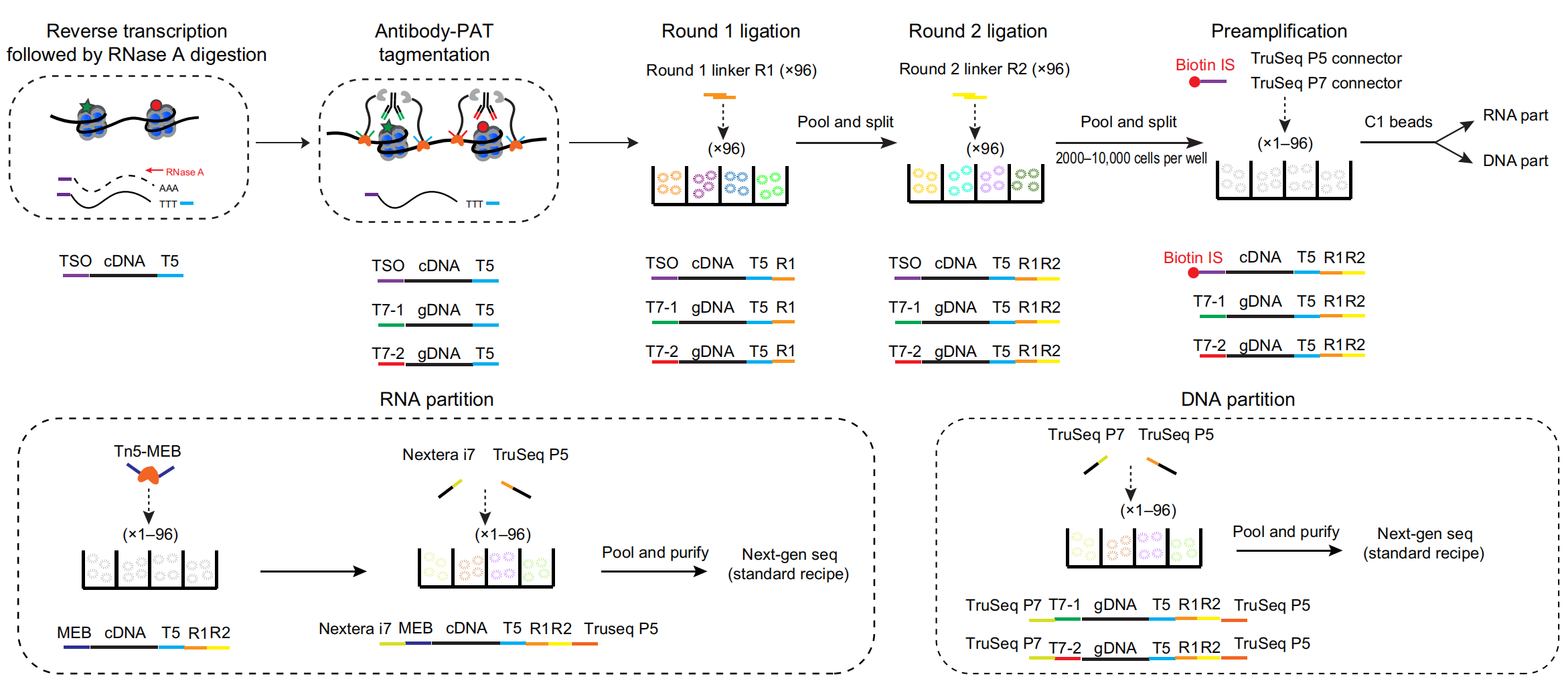
uCoTargetX
Single-cell joint profiling of multiple epigenetic proteins and gene transcription.
We developed uCoTarget, utilizing a split-pool barcoding strategy for realizing ultra-high throughput single-cell joint profiling of multiple epigenetic proteins. Furthermore, we developed uCoTargetX, an expansion of uCoTarget to simultaneously measure transcriptome and multiple epigenome targets. Together, our methods enable generalizable, versatile multi-modal profiles for reconstructing comprehensive epigenome and transcriptome landscapes and analyzing the regulatory interplay at the single-cell level. (Xiong et al. Science Advances, 2024)
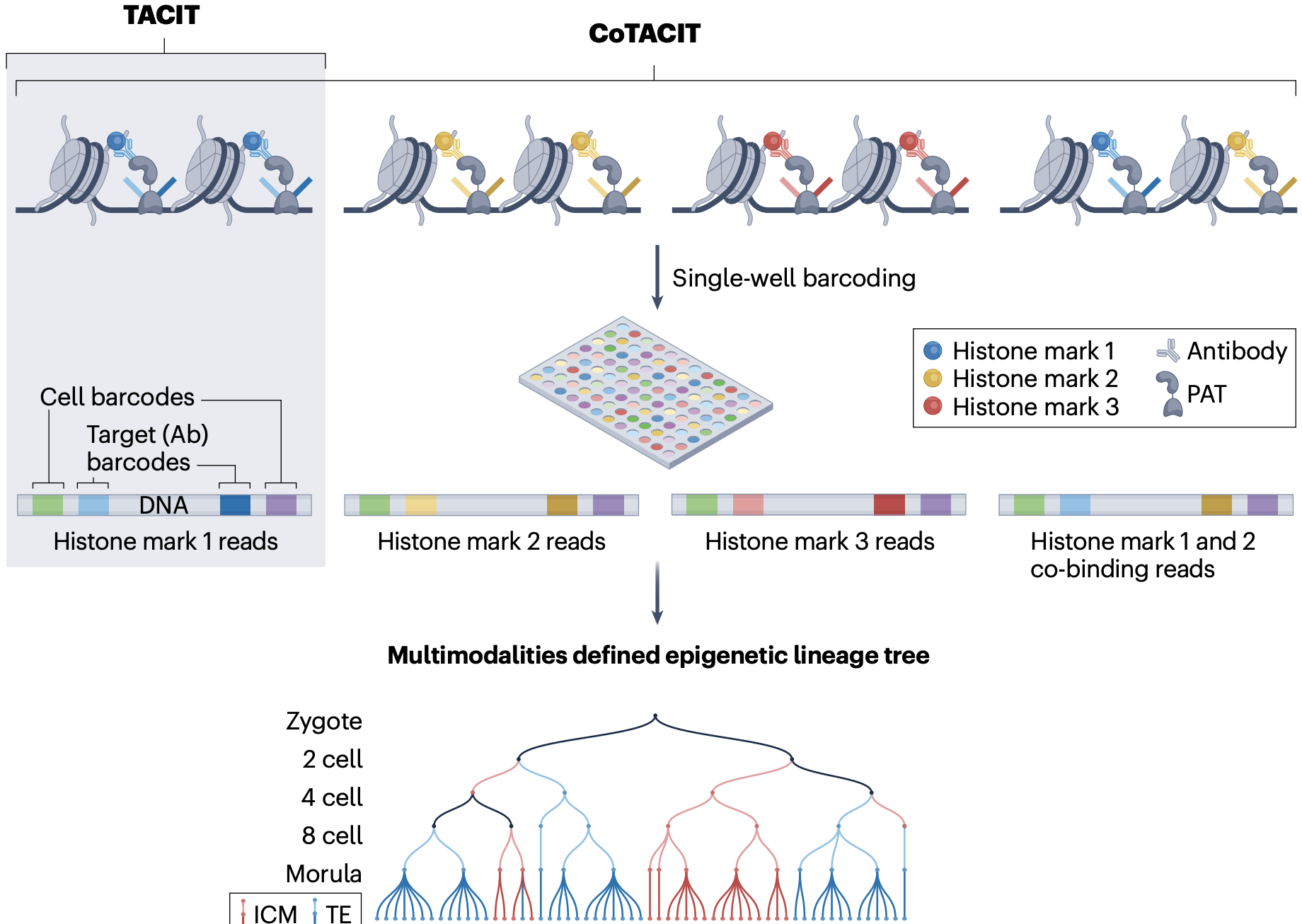
TACIT and CoTACIT
TACIT and CoTACIT for histone modification profiling in single cell and lineage tracing
We developed TACIT (target chromatin indexing and tagmentation) , single-cell chromatin indexing and tagmentation method that employs antibody-guided protein A–Tn5 transposition to barcode and map histone modifications genome-wide from as few as 20 cells.Built on TACIT, CoTACIT (combined assay of target chromatin indexing and tagmentation) simultaneously maps up to six histone modifications in the same cell. (Liu et al. Nature, 2025)