TECHNOLOGIES AND FUNDAMENTAL BIOLOGY
1. Single-cell omics technologies for probing complex gene regulation and cell fate.
Single-cell omics technologies for probing complex gene regulation and cell fate diversification. To achieve these goals, we have developed numerous novel single-cell technologies, including single-cell ChIP-seq (itChIP and CoBATCH) and CoTECH.
Single-cell itChIP-seq enabled genome-wide profiling of histone modifications and non-histone proteins from as few as 100 cells and various histone modifications of single-cell samples. Using sc-itChIP-seq to profile H3K27ac, we captured the earliest epigenetic priming along cell fate transition from naïve-to-primed pluripotency, and revealed the basis for cell-type specific enhancer usage during the differentiation of bipotent cardiac progenitor cells into endothelial cells and cardiomyocytes. Altogether, itChIP is a simple and effective ChIP-seq method for profiling chromatin states of epigenetically heterogeneous cell populations in many biological processes. (Ai et al., Nature Cell Biology, 2019)
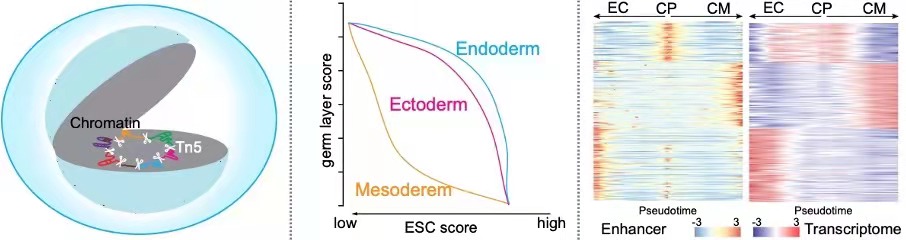
CoBATCH enables not only low-input epigenomic profiling in intact tissues, but also measure scalable up to tens of thousands of single cells per experiment. Through mapping of endothelial cell lineages from ten embryonic mouse organs through H3K27ac CoBATCH, we found that the epigenetic heterogeneity of ECs likely reflected their specialized functions and milieu-dependent developmental histories. Thus, CoBATCH is broadly applicable to understand the cis and trans modes of regulatory heterogeneity in physiological and pathological processes. (Wang et al., Molecular Cell, 2019)
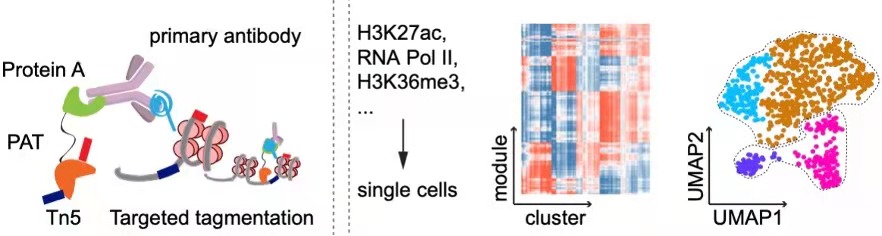
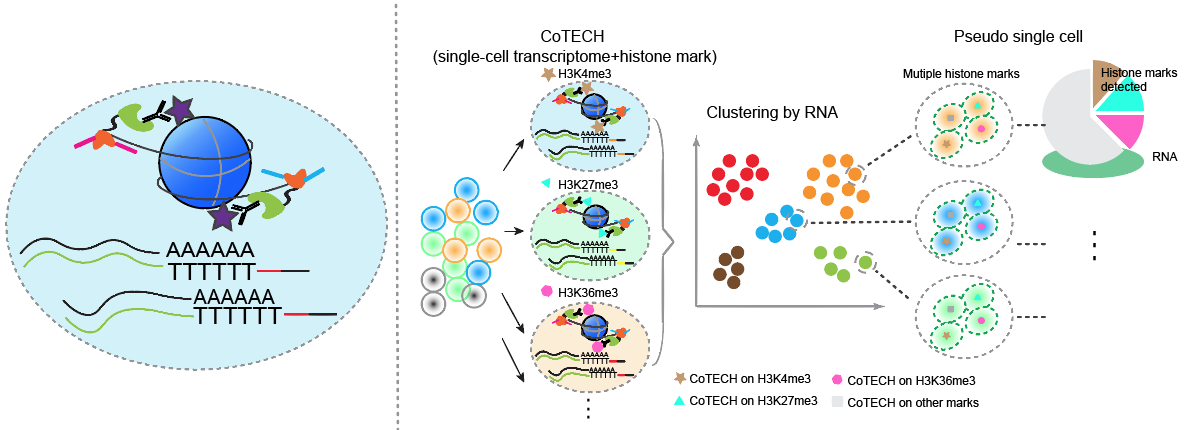
CoTECH is a joint method for profiling transcriptome and chromatin occupancy simultaneously in single-cells. We intergrated our previously developed CoBATCH together with modified single-cell RNA-seq method, to simutanously profile gene expression and chromatin binding in the same single cells. Furthermore, by aggregating data from cells with similar transcriptome (data from different experimental batches), we can obtain “pseudosingle-cells” that contain informations of several histone modifications or TFs binding, enabling higher-dimensional epigenomic reconstruction. (Xiong et al. Nature Methods, 2021)
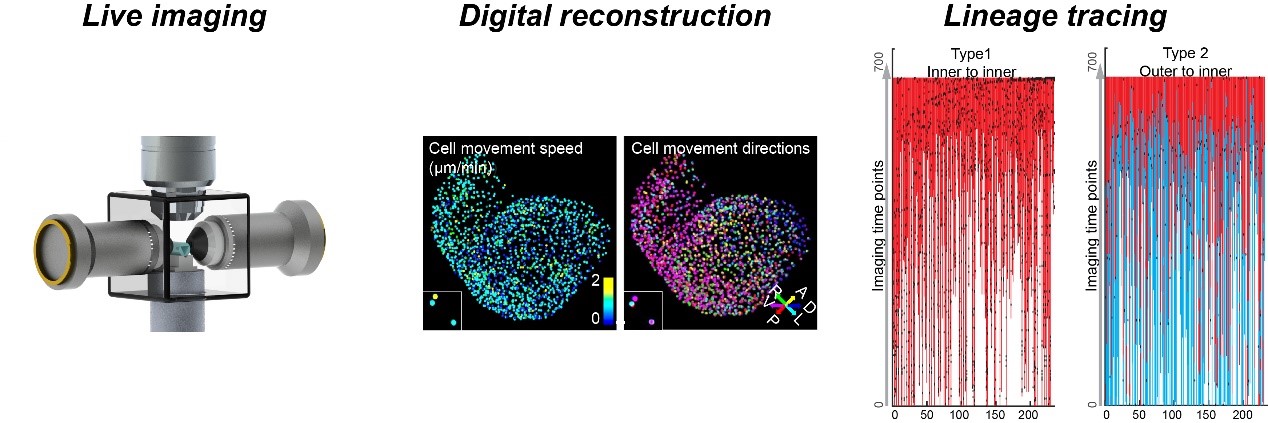
2. In toto live imaging for lineage reconstructions of organogenesis and regeneration.
A central theme in organogenesis is how cell lineages of divergent sources are spatiotemporally deployed to assemble complex organ structures in mammals. Our team overcame the technical challenges by optimizing mouse embryo culture and mounting methods, developing a vertical, dual-side illumination light-sheet microscope (vLSFM), equipping it with an integrated embryo culture module, together with a heartbeat-gated imaging module. With this integrative approach, we realized a 36-h, all cell-resolved imaging of developing mouse heart at 3-min intervals. Concepts derived include a new perspective for how dynamic cell behaviors are coordinated in the pivotal events of heart morphogenesis – ventricular chamber ballooning and trabeculation, challenging while unifying prevalent views based on classic population or single-cell snapshot analyses. Our work also paves the way for continuous live imaging of multi-scaled grand developmental schemes of organogenesis as well as subtle pathological alterations, e.g., in congenital heart disease models.
3. Cardiovascular development and regeneration.
Heart development requires precisely spatio-temporal specification of multiple cell lineages from early cardiac mesoderm. One major challenge in the field is to understand cardiac lineage plasticity, including identity establishment, specification and differentiation.
Through tackling the question of how epigenetic mechanisms coordinate postnatal heart maturation and terminally differentiated cardiomyocytes homeostasis, we have revealed that EED orchestration of heart maturation through interaction with histone deacetylases (HDACs) is H3K27me3 independent (Ai et al., Elife, 2017) . Interestingly, by inspecting the histone turnover in post-mitotic hearts, we identified EED augmented histone turnover to restrain enhancer over-activation through directly interacting with and engaging the BRG1-associated factor (BAF) complex for nucleosome exchange for stereotyped histone modifications from the free histone pool (Li et al., Circulation Research, 2019) . Taking adavantage of the heart developmental cues, we revealed the divergent requirements for EZH1 in heart development and regeneration. Specifically, EZH1 acted as a redundant catalytic subunit to EZH2 in depositing the repressive histone mark H3K27me3 to silence non-cardiac genes during heart development, while in regenerating neonatal heart after injury, EZH1 but not EZH2 “opened” chromatin landscape associated with reduced H3K27me3 and increased H3K4me3 at cell cycle relevant genes to promote neonatal heart regeneration after injury. (Ai et al., Circulation Research, 2017)
4. Combining single-cell omics and live imaging.
TECHNOLOGIES AND MEDICINE
1. Striving to apply our newly developed technologies to decipher cellular and molecular mechanisms in human diseases, in particular for various blood diseases, as well as diagnostic medicine
Leukemia is a blood cancer characterized by the accumulation of abnormal blood cells in the bone marrow. Past efforts in understanding the molecular basis of leukemia are limited by the lack of technologies, and the underlying mechanism of oncogenesis, drug resistance, and relapse are still remain unclear. Here, by using of newly developed single-cell epigenome profiling technologies-itChIP, CoBATCH and CoTECH, we are trying to unravel the molecular mechanism of leukemogenesis and relapse. Many projects are ongoing, some are listed as follows:
• Providing a comprehensive high-throughput mapping of histone modifications, TFs bindings, and RNA expression at multiple clinical time-point at single-cell level.
• Identification of the epigenome and transcriptome difference of leukemia stem cells (LSC) at several time-points: primary diagnosis, chemotherapy, and relapse after hematopoietic stem cell transplantation (HSCT).
• Uncovering specific epigenome features of LSC, including specific binding sites and specific modification types, which may serve as molecular targets for diagnostic medicine or target therapy.